Welcome to Suo Lab
We are a young research group at the Guangzhou Laboratory. If you are interested in working with us, please see more information on Positions.
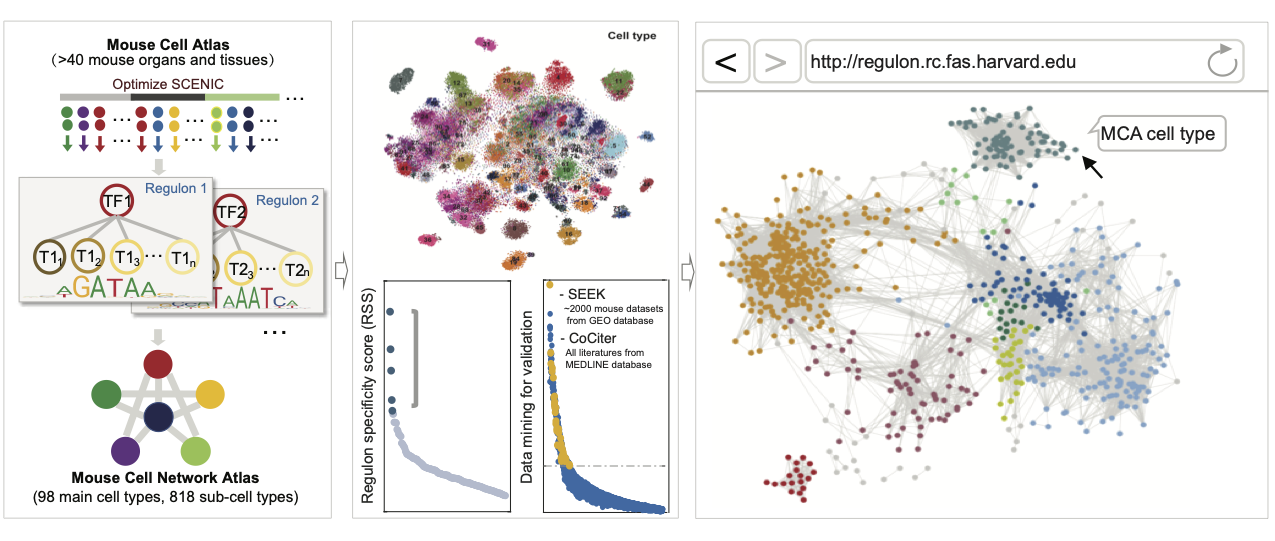
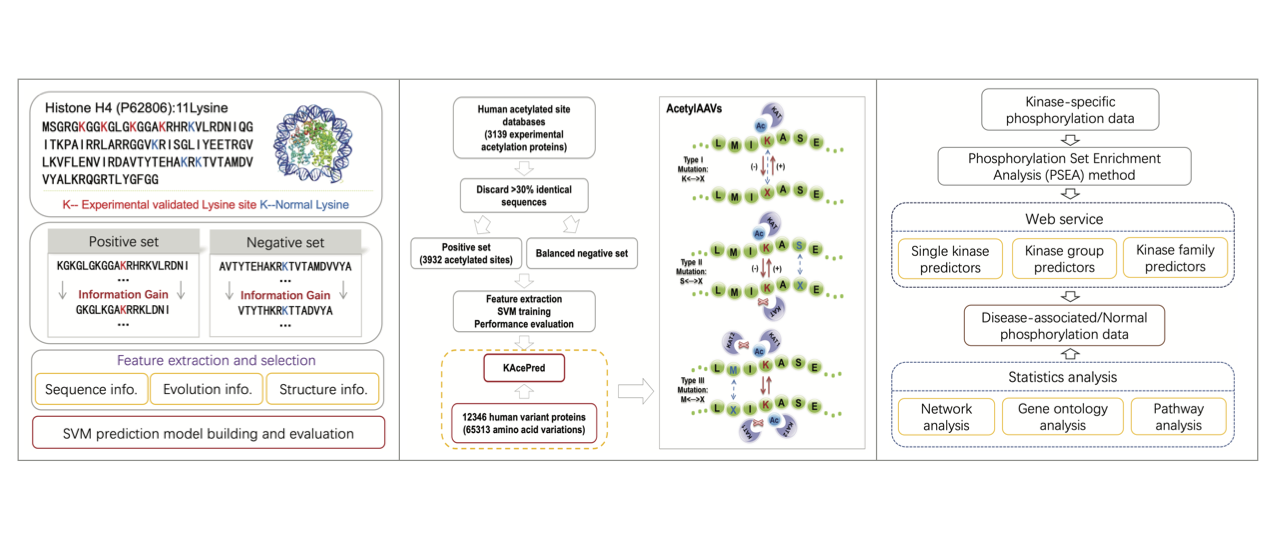
Transformative single-cell multi-omics and spatial technologies now provide an opportunity to dissect the state of individual cells at unprecedented resolution. Given the rapid development of different single-cell biotechnologies and new wave of big omics data with their own specificities in terms of complexity and information content, our aims is to use computational and theoretical approaches combined with multi-omics high-throughput sequencing data to quantitatively address the full-scale complexity of biological systems and derive novel biological insights. We are building an interdisciplinary team to tightly link computation and biological technologies, and gradually set up wet experiment systems for validating computationally inferred hypotheses. We will always be eager to seek collaborations, such as cancer immunology, within our rich scientific community to push forward our understandings of regulation mechanism and yield new therapeutic approaches for human disease treatment.
We are looking for passionate undergraduate/graduate student and postdoc to join the team (more info) !
News
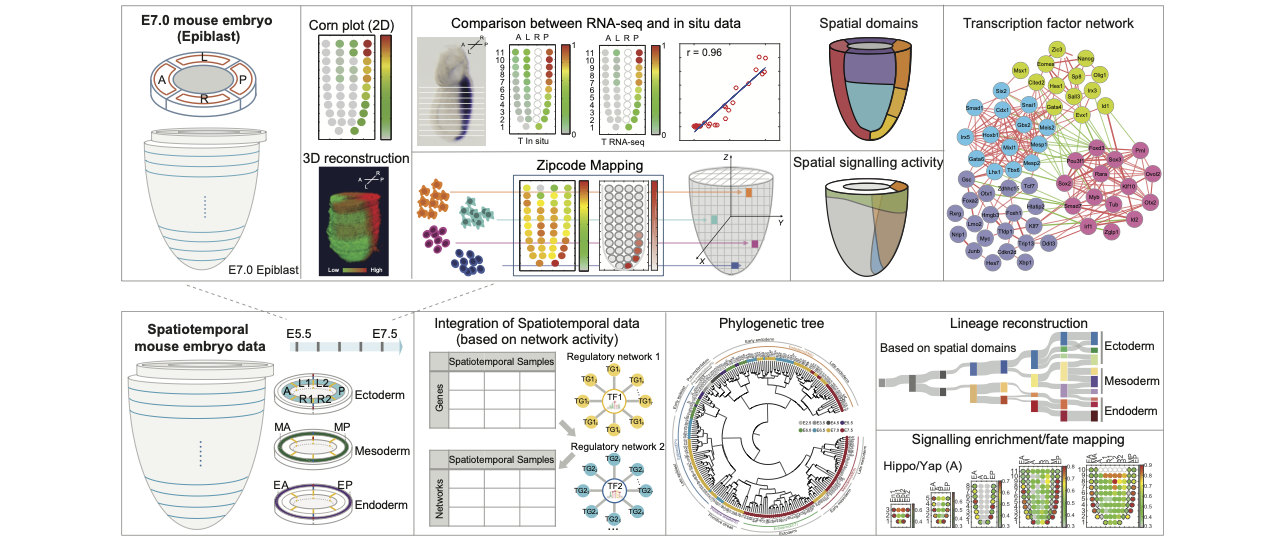
Jan 2026Our collaboration study of Integrated multi-omic atlas reveals the hierarchy of spatiotemporal regulatory networks of mouse gastrulation in Nature Communications, Congratulations to Bingbing!
Dec 2025Our collaboration study of Spatial and single-cell transcriptomics landscape of adenomyosis has been online publised in Journal of Advanced Research, Congratulations to Chunjie!
Nov 2025Our study of Assessment of computational methods in predicting TCR-epitope binding recognition has been online publised in Nature Methods, Congratulations to Yanping and Yuyan!
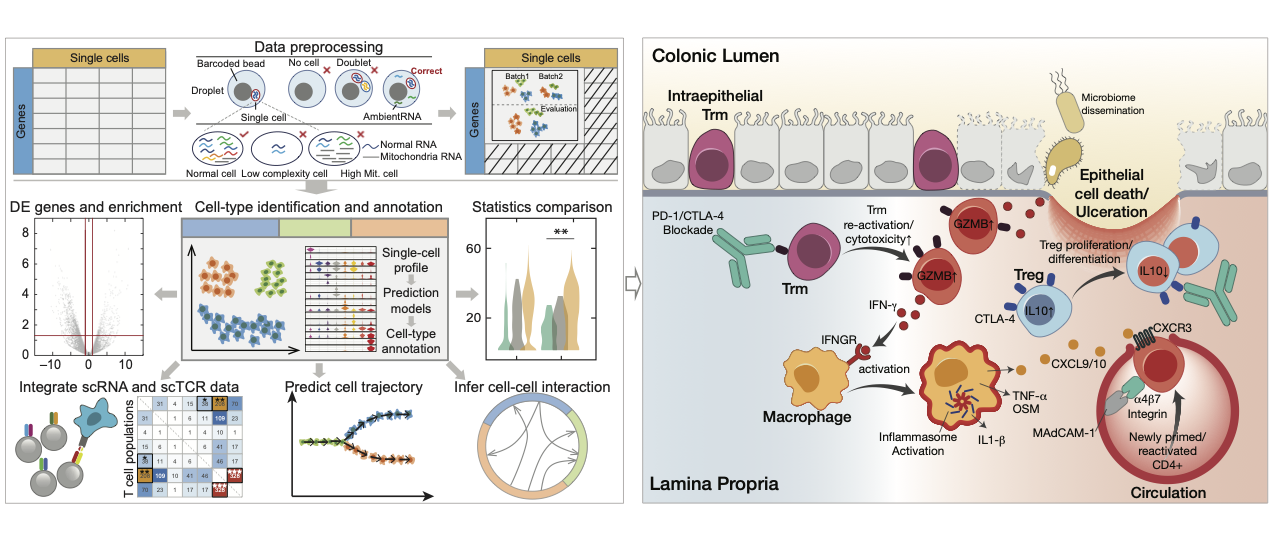
Nov 2025Our collaboration study of Uncovering the immune mechanisms underlying the emergence of immunotherapy-induced pneumonitis in lung cancer patients has been online publised in Nature Communications, Congratulations to Chunjie!
Jul 2025Our collaboration study of Human iPSC-based breast cancer model identifies S100P-dependent cancer stemness induced by BRCA1 mutation has been online publised in Science Advances, Congratulations to Jingxin!